Introduction
The promise of genomics in healthcare is massive—enabling personalized treatments, early disease detection, and better patient outcomes. But unlocking this potential isn’t just about sequencing genes—it’s about making that data actionable in real-time clinical workflows.
Traditional healthcare data systems weren’t built for genomics. They struggle with:
- Delays – Processing genomic data can take 5+ hours, delaying critical insights.
- Complexity – Genomic sequencing reveals millions of variants, yet EHRs capture only a handful.
- Data Silos – Inconsistent syncing between platforms like Salesforce, NetSuite, and EHRs creates trust issues.
- Scalability – Without a standardized framework, matching patient data to clinical insights is slow and inefficient.
That’s where FHIR Genomics Operations comes in—solving these challenges by making genomic data searchable, standardized, and usable at scale.
What is FHIR Genomics Operations?
FHIR Genomics Operations extends FHIR APIs to help healthcare organizations integrate genomic insights directly into patient care. It enables:
- Real-time risk scoring – Automatically updates patient risk levels as new genetic variants are discovered.
- Better clinical trial matching – Uses somatic variants to find eligible trials.
- Optimized medication dosing – Identifies pharmacogenomic insights to personalize treatment.
- Genomic screening – Flags hereditary conditions across generations.
- Improved data retrieval – Extracts genotype and phenotype insights not available in EHRs.
This eliminates the lag, inefficiencies, and inconsistencies that previously slowed genomic data adoption in healthcare.collection of variants. Given the growing number of pathogenic variants and the potential widespread adoption of polygenic risk scores in clinical practice, the complexity of re-analysis will continue to grow.
Furthermore, even when genomic data is standardized in FHIR format, variants can still be represented in different ways in different knowledgebases. This inconsistency makes it difficult to effectively search and aggregate genetic information, particularly when trying to match patient profiles against non-FHIR knowledge bases. These representation differences create extra complexity for developers building applications that use genomic data for clinical care, operations, or research purposes.
FHIR Genomic Operations
Various healthcare applications support use cases that rely on complex genomics data; these applications require capabilities that:
- update a patient’s risk as new pathogenic variants are incorporated into clinical practice
- extract a specific genotype or phenotype information for a patient or population in a form or format that does not already exist on the EHR server or in a FHIR format
- screen the patient and parents for actionable hereditary conditions
- match a cancer patient to available clinical trials based on identified somatic variants
- identify risk for adverse medical reactions based on pharmacogenomics variants or optimizing the dose of a medication
FHIR Genomics Operations is introduced to support these and other use cases. ‘Operations’ is a FHIR construct that extends RESTful FHIR API’s CRUD (Create/Read/Update/Delete) actions in a standardized way so that the defined use cases implemented via the EHR can be enabled through standardized responses generated using a server. The FHIR Genomics Operations encompasses the systems, processes, and infrastructure needed to manage and integrate genomic data in healthcare settings to allow for various use cases.
FHIR Genomics Operations was developed by drawing inspiration from the architecture that enabled seamless sharing of radiological images between healthcare providers. This sharing is facilitated through a Picture Archiving and Communication System (PACS) and Digital Imaging and Communications in Medicine (DICOM) standards. PACS stores image data that cannot be housed within the Electronic Health Record (EHR), while DICOM sets the standards for storing and transmitting images.
In a similar way, the Genomics Archiving and Communication System (GACS) functions as a genomics data server outside of the EHR. Located within healthcare organizations, this server stores raw genomic sequence data received from sequencing laboratories, as well as other reference data. The GACS integrates with applications via APIs based on FHIR or FHIR Genomics Operations.
The goal of FHIR Genomics Operations is to support a wide range of applications, including updating a patient’s risk score, optimizing medication dosage, identifying the risk of adverse drug reactions based on pharmacogenomic variants, matching patients to clinical trials based on somatic variants, and screening for actionable hereditary conditions. To achieve this, the system must bioinformatically process and calculate data from numerous variant representations stored in both data repositories and knowledge sources. Genomic operations that normalize the data store simplify the complexity of managing this information by providing a single, unified representation of genetic variation. This allows both industry and academic developers to focus on creating high-performance, user-friendly, and scalable Apps. In summary, FHIR Genomics Operations serves as the abstraction layer, providing a uniform interface to various applications, regardless of the underlying data structures.
FHIR Genomics Operations has been categorized along two orthogonal axes – subject vs population and genotype vs phenotype. The Operations will allow an application to retrieve genotypic or phenotypic characteristics of a specific subject or retrieve a number or list of patients with a specific genotypic or phenotypic disposition.
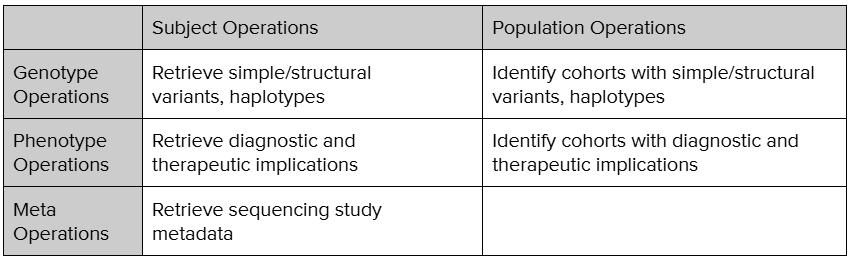
The find-subject-variant is a ‘Subject’ and ‘Genotype’ operation. This operation is designed to retrieve a single variant for the given subject. Meanwhile, find-population-tx-implication is a ‘Population’ and ‘Phenotype’ operation, where the operation is designed to retrieve a cohort of patients with specific phenotypic characteristics, like moderate metabolizers of clopidrogel. As the name indicates, meta Operations retrieves the metadata about the genomics study. The development of around 15 such Operations are being evaluated. More operations will continue to be added as the use cases are developed further.
Genomic Liftover
All FHIR Genomics Operations allow for a genomic region-based searching and the expectation of server-side normalization and genomic liftover. These processes can be highly resource-intensive due to the complexity of variation representation. To provide a consistent query approach for variants, operations include a “ranges” query parameter. For example, to query variants in the APC gene, one can use the “find-subject-variants” operation with the parameter ranges= “NC_000005.10:112707497-112846239”; these coordinates are for the build 38. The coordinates for build 37 are “NC_000005.9:112043194-112181936”. The initial coordinates were provided for the Build 38 range; here, it will be necessary for the server to ‘liftover’ (available solutions) the query region into the Build 37 coordinates and identify the variants against those coordinates. NCBI’s resources serve as the definitive reference for the location of genes based on the human genome.
Variant Normalization
Variants can be represented in different ways, and against different references. The following HGVS and VCF representations refer to the same variant:
- NM_001195798.2:c.12G>A
- NM_001195803.2:c.12G>A
- NC_000019.9:g.11200236G>A
- NC_000019.10:g.11089560G>A
- NC_000019.10:11089559: G: A
It is expected that FHIR Genomics Operations returns the variants in the requested regions irrespective of the format in which these variants are stored in the database. There are several standard databases that store information about variants and their implications in human health; these include COSMIC, PharmGkb, ClinVar, and CIViC. It is expected that the normalization operations against these or other knowledge databases is performed at the server side. One approach for this is to transform these different representations into the canonical SPDI format. The queries can then be bounced off this format. NCBI’s variation services provide APIs that can be used to normalize variants into the SPDI format.
Kipi.ai’s Role in Driving Genomic Innovation
At Kipi.ai, we’re building the future of healthcare data management with our Health DataHub, leveraging Snowflake’s native capabilities to:
- Streamline FHIR data ingestion & transformation
- Enable rapid AI-driven analysis with FHIR Server Enablers
- Simplify real-time data access using Streamlit Apps
With Kipi.ai + Snowflake, healthcare organizations can unlock new frontiers in genomic research, precision medicine, and clinical decision support.
Conclusion
FHIR Genomics Operations is helping turn raw genomic data into real-world clinical insights. By standardizing data access, variant normalization, and interoperability, these innovations will drive faster diagnoses, smarter treatments, and better patient outcomes.
As genomic applications grow, so will FHIR Genomics Operations—transforming healthcare one breakthrough at a time.
About kipi.ai
Kipi.ai is a leading analytics and AI services provider, specializing in transforming data into actionable insights through advanced analytics, AI, and machine learning. As an Elite Snowflake Partner, we are committed to helping organizations optimize their data strategies, migrate to the cloud, and unlock the full potential of their data. Our deep expertise in the Snowflake AI Data Cloud enables us to drive seamless data migration, enhanced data governance, and scalable analytics solutions tailored to your business needs. At kipi.ai, we empower clients across industries to accelerate their data-driven transformation and achieve unprecedented business outcomes







